Masterclass Series
Spatial Biology – image analysis and practical application
#VisiopharmSpatialSeries
We have completed our final Spatial Biology Masterclass in 2024, but we’ll be back in 2025 with more exciting topics and expert speakers, so stay tuned!
In the meantime, feel free to watch the completed classes on demand. Full list below.
Spatial biology has undergone a significant evolution, transitioning from traditional histopathologic morphology methods to modern multi-omic approaches, largely due to technological advancements. Understanding the entire tissue context, from mapping tissue to elucidating expression profiles within each cell or cellular compartment, serves as a crucial precursor to gaining deeper insights into the pertinent participants of disease progression.
While immuno-oncology has prominently adopted multiplexed spatial biology techniques, other fields such as neuroscience, dermatology, hepatology, animal models and others are also recognizing the potential of spatial biology methodologies.
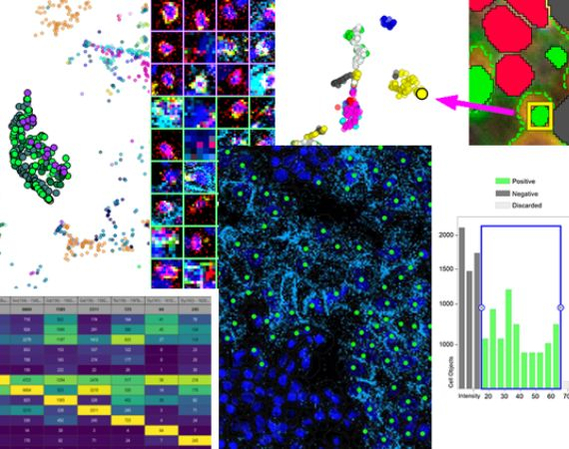
Given the complexity of data these methodologies produce, there's a critical need for specialized tools. Visiopharm's Phenoplex platform offers a complete multiplex image analysis workflow that saves you time and instils confidence in the accuracy of your results.
Join this masterclass series led by experts who will address all facets of spatial biology. Participants will gain insights into cutting-edge methodologies, techniques, and tools essential for navigating the intricate spatial biology landscape effectively.
Topics to be covered in the series
More topics will be added on an ongoing basis.
- Spatial biology 101
- Reliability of results
- The importance of tissue mapping and cell segmentation
- Multimodal analyses combining -OMICS
- Neighborhood analysis
- And much more...
We are excited to announce that this Masterclass series will be a mix of both in-person and virtual events.

Dr Fabian Schneider
Product Manager Research, Visiopharm
17 April, Wednesday, 4pm CEST/10am EST
Mapping the biological landscape: Turning multiplex images into knowledge
Abstract
Spatial biology is revolutionizing molecular biology by enabling the examination of cells within their tissue microenvironment. It empowers scientists to reconstruct tissue structures and compartments and investigate intricate cell-to-cell interactions and dependencies. This level of detail is unparalleled by any other available method. The resulting multiplexed images require easy-to-use image analysis tools that are tailored specifically for biologists and pathologists. In this masterclass, we illuminate the significance of spatial biology in modern research, unveiling its potential to revolutionize our understanding of biological processes and disease states.
We will discuss its capacity to unveil hidden biological nuances and disease mechanisms within tissue landscapes. Emphasizing the practical needs of biologists and pathologists, we will delve into:
- Image visualization and quality control.
- Tissue segmentation strategies to delineate anatomical regions and tissue compartments accurately.
- Cell segmentation, both turn-key and enhanced.
- Phenotyping for characterizing cellular populations and discerning pathological features.
- Spatial neighborhood analysis tools, illuminating the spatial relationships vital for understanding microenvironmental influences.
- The demand for interactive data interrogation platforms, facilitating intuitive exploration of results and hypothesis generation.
Join us as we navigate the complexities of spatial biology and multiplex image analysis, empowering biologists, and pathologists to decode the mysteries concealed within biological landscapes. By bridging the gap between images and insights, Phenoplex equips researchers with the tools to navigate and understand the complexities of biological systems with precision and clarity.
Biography
Dr Fabian Schneider is part of Visiopharm’s R&D and Product Management team, responsible for phenotyping products as well as service projects for custom APP development. Fabian has over 10 years of international experience in cancer biology and immuno-oncology, working in academic research labs, clinical research teams and computational pathology groups in both academia and biopharma.
Fabian received his Dr phil. nat. in Cell Biology in 2011 from the Johan Wolfgang Goethe University Frankfurt, Germany.

Karen McClymont (left) / Image Analysis Project Manager and Gabriel Reines March (right) / R&D Project Manager, OracleBio
15 May, Wednesday, 4pm CEST/10am EST
Using Visiopharm to support image analysis workflow QC and trust in data
Abstract
Delivering high-quality, quantitative data from digital pathology images is one of OracleBio’s core values. Tools such as Visiopharm enable us to achieve this by building robust, AI-powered image analysis workflows.
In this masterclass, we explore the QC strategies used to ensure accuracy and reproducibility in the following case studies:
- The creation of a pathologist-validated and scalable image analysis workflow for a retrospective clinical study involving over 2,200 IHC-stained colorectal polyp whole slide images.
- The use of Deep Learning apps to segment morphologically distinct cell subpopulations and the Phenoplex workflow to streamline the thresholding process for spatial phenotyping in multiplex IF-stained pre-clinical tissue.
Biography
Karen McClymont
With a PhD in Biochemistry and her active involvement and support in the analysis of some of OracleBio’s most complex studies, Karen plays a key role leading OracleBio’s image analysis projects and overseeing the team’s continued development.
Gabriel Reines March
With a PhD in Biomedical Image Processing and a background in Electrical Engineering, Gabriel is the OracleBio R&D team lead. He oversees and manages the group’s project pipeline and ensures that the company stays at the bleeding edge of the industry.
Gabriel also manages OracleBio’s involvement in the INCISE project – a collaborative effort between industry, academia and the UK’s National Health Service.

Professor, M.D. Anderson
19 June, Wednesday, 4pm CEST/10am EST
Multiplex Multi-Omics - The Future of Spatial Biology
Abstract
The future of spatial biology promises to reveal the intricate functional interplay between cells, a crucial aspect for understanding disease progression and therapeutic targeting. To decode these cellular conversations, a multiplex multi-omics approach is essential, enabling comprehensive identification and comprehension of diverse analytes. Presently, our capability is limited to combining a few analyte classes on a single slide, such as proteins and mRNA, often at the expense of one class. We can be creative in our assays to go beyond, which reveals the challenge in integrating informatics to not only unify these diverse data types but also to cross-validate and interpret the interactions and their effects on multiple cells in real-time. As cells continually move and interact with different or novel groups, understanding how one cell influences another, and how this influence propagates, is critical. Advancing these techniques will be pivotal in elucidating the dynamic cellular dialogues that underpin disease mechanisms and therapeutic responses.
Biography
Jared started his carrier at Texas A&M University learning about patterns in genes and proteins, allowing and facilitating subcellular protein trafficking. As he has progressed to MD Anderson Cancer Center, he has scaled to cellular trafficking attempting to understand the spatial distribution of cells in organ systems during disease. As in many parts of life, form equals functions. How our cells organize speaks to how they function and respond to their local environment. Bringing together multi-omics approaches allows for greater clarity in these imaging snapshots that are collected.

Janusz Franco-Barraza. MD, PhD
Research Assistant Professor at Cukierman Lab & Manager at Spatial Immuno-Proteomics Facility, Fox Chase Cancer Center
17 July, Wednesday, 4pm CEST/10am EST
HOST-Factor: Unveiling pancreatic cancer microenvironment neighborhoods through highplex imaging and spatial profiling
Abstract
Solid tumors' complexity extends beyond the genetic and functional diversity of cancer cells to include the tumor microenvironment (TME). Understanding the TME's complexity requires a comprehensive analysis of the composition, functional states, and spatial distribution of its cellular components.
In this session, we will explore the application of basic research discoveries through the Harmonic Output of Stromal Traits (HOST) for the evaluation of the TME. HOST is designed to identify TME cells, while the HOST-Factor quantifies their functional states. The HOST-Factor is a numerical value reflecting the relative contribution of cancer-associated fibroblasts (CAFs) to tumor-suppressive or tumor-promoting functions.
Our workflow combines automated high-plex immunofluorescent microscopy with AI-guided image analysis to generate single-cell HOST-Factor values. This approach, applicable to the entire TME or specific regions of interest, provides spatial distribution data and identifies potential tumor-promoting or suppressive neighborhoods.
This talk will highlight the application of Visiopharm’s Phenoplex workflow and its Neighbor Counts module to identify and evaluate HOST+ CAF populations and their spatial distribution. Developed specifically for a pancreatic cancer clinical trial at Fox Chase Cancer Center, this user-friendly pipeline is flexible and useful for assessing diverse stromal compartments in distinct solid tumor cases.
Biography
Dr. Franco-Barraza obtained his Ph.D. in Molecular Biomedicine in 2010 from CINVESTAV-IPN (Center for Research and Advanced Studies of the National Polytechnic Institute, Mexico). During his postdoctoral work at Fox Chase Cancer Center, he studied integrin-based activation mechanisms used by cancer-associated fibroblasts (CAF) within their self-secreted extracellular matrix (ECM), acting as complex entities called CAF units. He focuses on understanding the functional traits of these CAF units and their roles in tumor promotion or suppression.
Utilizing approaches including multiplexed immunofluorescence and artificial intelligence (AI)-driven digital image mining, he is dedicated to understanding the prevalence and interplay of CAF units with other resident cells within the tumor microenvironment. Currently, his research aims to profile the stroma of cancer patients to validate and implement fibroblastic biomarker signatures that could predict treatment responses and disease outcomes.

Bora Gurel, MD, PhD
Clinician Scientist / Pathologist at The Institute of Cancer Research
26 September, Thursday, 4pm CEST/10am EST
Hyperplex IF analysis for tumour immunology: Our experience with Visiopharm
Abstract
Prostate cancer remains a significant health challenge globally, with metastatic castration-resistant prostate cancer (mCRPC) marking a critical juncture in patient care. The complex interactions between tumour and immune cells within the tumour microenvironment are pivotal for disease progression and therapeutic outcomes. In this presentation, I share our experience with using hyperplex immunofluorescence (IF) analysis with Visiopharm software to explore these interactions. I describe our approach to tissue segmentation, the determination of positivity in IF images, and the phenotyping of various cell populations. Moreover, I will discuss our observations on the spatial distribution of immune cells within the tumour microenvironment and its implications for prognosis. This work highlights the utility of hyperplex IF analysis in deepening our understanding of the immune landscape in mCRPC, offering new perspectives on prognosis and potential therapeutic avenues.
Biography
Dr. Gurel is a computational pathologist specialising in urogenital pathology with extensive experience in computerized image analysis, deep learning, molecular pathology, and cancer research. He is currently working as a clinician scientist/pathologist with the De Bono team at the Institute of Cancer Research, London.
The on-demand recording for this Masterclass will be made available as soon as the data has been published.

James Pemberton, PhD
Product Application Scientist at Standard BioTools Inc.
30 October, Wednesday, 4pm CET/10am EST
New Views in Spatial Biology: Multiplexed Imaging for Rapid Whole Slide Analysis
Abstract
Join us for an insightful seminar on the Hyperion XTi™ Imaging System and its pivotal role in advancing spatial biology. This emerging field is transforming our understanding of tissue organization in health and disease, with implications for predicting immune and therapeutic responses, improving drug development strategies, and potentially preventing or curing diseases.
While conventional microscopy techniques use fluorescence to visualize protein or RNA targets in tissue samples, they face challenges due to spectral overlap and tissue autofluorescence. The Hyperion™ XTi overcomes these limitations by utilizing Imaging Mass Cytometry™ (IMC™), a fluorescence-free approach enabling detection of 40+ targets.
IMC has expanded its capabilities with new whole slide imaging modes, which include Preview Mode, Cell Mode and Tissue Mode. These modes facilitate rapid and detailed analysis, supporting automated, continuous imaging of more than 40 large tissue samples (400 mm²) weekly. This significantly accelerates researchers' ability to gain insights from complex biological samples.
Phenoplex™ software from Visiopharm® complements this hardware by offering a comprehensive workflow for data analysis, including automated ROI selection, phenotyping and spatial analyses of IMC images. This seminar will demonstrate how the multi-modal features of Hyperion XTi, combined with advanced analysis tools from Visiopharm, are revolutionizing our approach to spatial biology and opening new avenues for biological assessment.
Biography
James holds a PhD in medical biophysics from the University of Toronto, specializing in advanced fluorescence microscopy techniques, cell biology and cell signaling mechanisms. At Standard BioTools, he currently works within product management and collaborates with R&D teams to leverage the multi-plex capabilities of the Hyperion XTi Imaging System. His goal is to drive innovation in spatial biology, enhancing our understanding of complex cellular interactions and tissue architecture in both health and disease.

Rachel Pennie, MSc
Scientific Officer at CRUK Scotland Institute (formerly Beatson Institute)
20 November, Wednesday, 4pm CET/10am EST
A novel global phenotyping method using artificial intelligence to overcome the cell segmentation bottleneck
Abstract
Multiplex methods for the detection of protein expression generate extremely data-rich images of intact tissue sections. These images are invaluable for the quantification and analysis of complex biology and have huge potential for biomarker development. However, their interpretation presents a considerable data analysis challenge which limits their utility, especially in clinical workflows.
The key task from these images is often cellular phenotyping - e.g. the identification of immunophenotypes, or the separation cells with compartment-specific protein expression from others. Most current automated methods depend upon a cellular segmentation step to support this, and segmentation has become an area of intensive research in AI (Artificial Intelligence), but the task is very difficult and no universally accepted method has been identified.
Therefore, methods of cellular phenotyping which operate without the need for segmentation would be very valuable. To meet this challenge, we have developed an AI cellular phenotyping method which does not require segmentation at all.
We use the U-Net backbone within the Visiopharm deep learning module to train our algorithm using expert human annotations of entire cellular regions. Crucially, we need only train for a single example of each compartmental stain (nuclear/cytoplasmic/membranous), and the algorithm then assigns class identities to cells without the need for segmentation of nuclei cytoplasm or membranes, using regional information in a manner analogous to a human expert, who may not need to be confident about cellular boundaries to accurately phenotype a cell.
The method is comparable in accuracy to segmentation-based methods, is highly transferrable, and represents a highly novel approach to quantitative image analysis with the potential to radically alter the way that we analyse these data-rich images.
Biography
Rachel Pennie MSc is a translational histologist in the deep phenotyping advanced technology facility within the Le Quesne Lab at the CRUK Scotland Institute. She is an expert in the development and application of Visiopharm deep learning methods to multiplex and high plex histopathological images.

Clive Taylor
Emeritus Professor at Keck School of Medicine, USA

Dr Rasmus Røge, MD, PhD
Hematopathologist and Clinical Associate Professor, Aalborg University
Dr Rasmus Røge is a pathologist and Clinical Associate Professor affiliated with Aalborg University. His daily work focuses on hematopathology, specializing in the study and diagnosis of blood-related disorders. Dr Røge has been actively involved in the Nordic Immunohistochemical Quality Control (NordiQC) since 2012, an international external quality assessment scheme that serves diagnostic pathology laboratories worldwide. With more than 30 publications, his research primarily centers around quality assessment, methodology in Immunohistochemistry and introduction, validation and optimization of new biomarkers in pathology.

Prof Ralf Huss
BioM Biotech Cluster Development GmbH & University Hospital Augsburg, Germany
Ralf Huss is a Professor of Pathology and currently the Managing Director and CEO of the Biotechnology Development Agency in Munich, Germany. Prior to this role, he was the founding director of the Institute for Digital Medicine at the University Hospital Augsburg, Germany. Dr. Huss is board-certified in anatomical, experimental, and molecular pathology, with over 30 years of experience in international academic institutions and the pharmaceutical industry with a focus on histopathology, immunology, cancer research, and digital medicine.

Omar Baba
MD, Clinical Pathologist, Henry Ford Hospital, USA
Omar Z. Baba, M.D. is a Pathology Informatics Fellow at Henry Ford Health, Detroit, mentored by Dr J Mark Tuthill. A Clinical Pathologist trained at the American University of Beirut Medical Center in Lebanon, he has been at the forefront of pathology informatics, deeply engaged in multiple informatics operations at the department of Pathology and Lab medicine at HFH and notably leading a validation initiated on an AI-driven image analysis tool, which he presented at the 2023 PI Summit in Pittsburgh.

Nils 't Hart
Pathologist, Isala Hospital, The Netherlands

Paul J van Diest
Professor, UMC Utrecht, The Netherlands

Colin Tristram
CEO, Histocyte, UK

Steve Bogen
CEO, Boston Cell Standards, USA

Regan Fulton
CEO, Array Science, USA
Dr Fulton received his MD and PhD from the University of Minnesota and completed his residency in Anatomic Pathology at Stanford University. Following residency, he completed fellowships in Surgical Pathology and Immunodiagnosis at Stanford University and is board-certified in Anatomic Pathology. He is the founder and CEO of Array Science, LLC, a manufacturer of control and proficiency-testing material. He holds multiple patents for making tissue and cell culture microarrays. He now works full-time at Array Science, while providing pathology support in the development of diagnostics, as well as various phases of clinical trials. Dr Fulton has served as a consultant and paid speaker for several pharmaceutical and biotechnology companies.

Michael Grunkin
CEO, Visiopharm, Denmark

Dirk Vossen
CDO, Visiopharm, the Netherlands
Dirk Vossen leads a cross-functional team to develop diagnostic and clinical applications of digital pathology. His track record of creating value through innovation in digital and computational pathology spans the entire range of development, from ideation through validation and certification of medical devices, as well as commercialization strategies.
Posters being presented at AACR
Session Date and Time: Sunday Apr 16, 2023 1:30 PM - 5:00 PM
Published Abstract Number: 619
Poster Board Number: 20
Poster Section: 21
Authors: Y. Shi et al. from Xi'an Jiaotong University, China,; with MD Anderson Cancer Center
Session Date and Time: Sunday Apr 16, 2023 1:30 PM - 5:00 PM
Published Abstract Number: 1018
Poster Board Number: 28
Poster Section: 41
Authors: D. Zielinski et al. from Discovery Life Sciences
Session Date and Time: Monday Apr 17, 2023 1:30 PM - 5:00 PM
Published Abstract Number: 2322
Poster Board Number: 3
Poster Section: 45
Authors: B. Dennison et al. from Lanterne Dx
Session Date and Time: Tuesday Apr 18, 2023 1:30 PM - 5:00 PM
Published Abstract Number: 3588
Poster Board Number: 13
Poster Section: 3
Authors: M. Pore et al. from NCI Frederick, MD
Session Date and Time: Tuesday Apr 18, 2023 1:30 PM - 5:00 PM
Published Abstract Number: 4625
Poster Board Number: 15
Poster Section: 4
Authors: B. O’Neill et al. from Visiopharm; with Standard Bio Tools